28-Feb-2023
Norbert Vischer
Bacterial Cell Biology
University of Amsterdam
Download SporeTrackerX
Documentation (pdf)
Other links:
Nikon .nd2 Import
Embedded Macros on GIT
Shadow Directory
SporeTrackerX is an ImageJ/ObjectJ project for the analysis of germintion and outgrowth of bacterial spores.
Due to consequent usage of virtual stacks, it allows to browse through very large time-lapse data sets (TByte) that are stored on external harddisks connected to a normal desktop or laptop computer.
In addition to provide the SproreTrackerX software, the documentation (pdf) proposes a suitable file structure and explains the conversion of Nikon .nd2 to TIFF files, the analysis of bacterial germination and growth, and the usage of suitable hardware.
Important parameters are doubling time TD, start of germination time, and burst time.
SporeTrackerX is a follow-up program of SporeTracker and accepts multi-channel hyperstacks. However, only the first channel (phase-contrast) is evaluated in this example.
Features
- Handling and converting Nikon .nd2 file sets up to 1 TB.
- Preparing a file structure
- Analysis of germination and bacterial growth

Choose menu ObjectJ>Analyze Spores
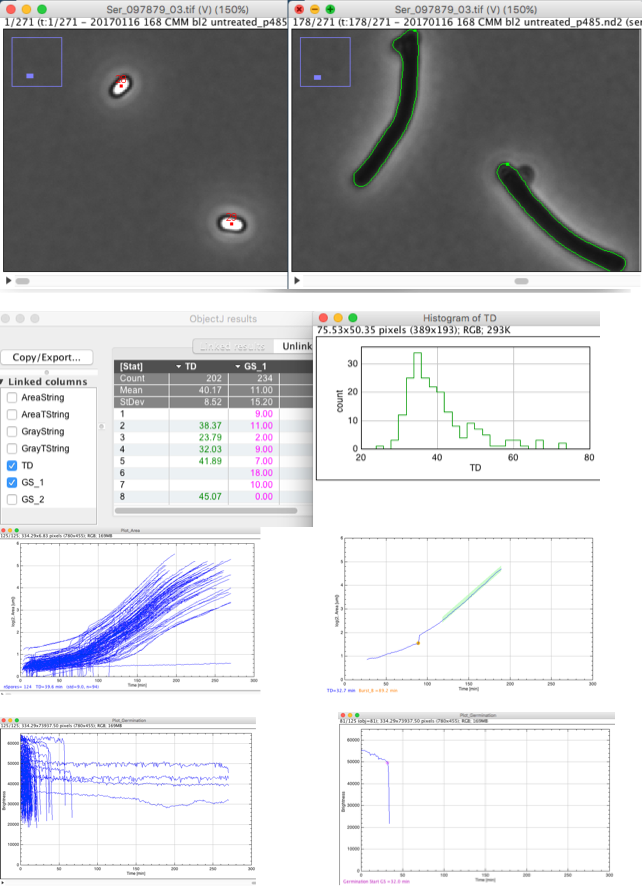
Spores will be marked with red points until germination and with green contours after germination.
Click OK if asked to switch to Navigation Mode
These commands are defined by the embedded macros of SporeTrackerX:
Bright and dark particles will be detected throughout the movie and identified as growing objects. An "object" corresponds to the history of a spore throughout the movie, and is marked with a red "Bright" marker per frame before germination, and with a green "Contour" marker per frame after that as long as it can be traced.
The number label appears in frame#1. Per spore object, the growing area versus time is recorded and stored as string in ObjectJ Results column "AreaPairs". Such a string consists of value pairs separated by semicolon, and each pair is consists of a time and area value separated by a space. This same technique is used for "GrayPairs" to quantify germination (bright to dark transition). After analysis, "PlotGrowth" and "PlotGermination" are created from the corresponding columns.
"PlotGrowth" and "PlotGermination" are created from the corresponding columns. A plot window is a stack holding one plot per object. One additional frame is appended to the stack containing a superimposition of all plots. Typically, a plot window needs not toe be saved, as it can be rebuilt at any time.
Opens all linked images as virual stacks, i.e. only a single frame is loaded into RAM. The large stacks should be virutally open so that many operations such as double-clicking a "linked image" or a "linked result" does not loose time loading the entire stack.
Qualifies/disqualifies the object shown in the current growth plot, sets the plot background to white/ gray, and sets the value in column "Q" to one/zero.
(Dis-)qualifies all objects depending on value in column "Q", and provides the option to kill unqualified objects (which will re-draw the plot windows).
Shows project path, long and short file names, and batch number. Long file names derived from the label in the stack's metadata.
Use this by activating the Plot_Growth window, position the cursor upon the time where the burst should be marked, and press the shortcut key "B". Any old burst marking will be replaced by the new one, or removed if key was pressed twice on same position.
Similar to "Sign Burst", but intended to mark the first division.
Similar to "Sign Burst", but sets the starting point of the green line whose slope defines doubling time TD.
Similar to "Sign Burst", but sets the end point of the green line whose slope defines doubling time TD.
Arranges windows "PlotGrowth" and "PlotGermination" on the right hand of the screen, and activates the navigation tool in ObjectJ Tools ("N" icon). If an object was selected (e.g. clicked in the ObjectJ results row), the Navigate command activates the plot of this object in both plot stacks. While the "N" tool in ObjectJ Tools is active, the user can click and drag along the time axis of the plot in order to play-back the movie synchronously.