| Author: | Norbert Vischer (vischer (at) uva . nl) |
| History: |
2017-07-24: Version 17 (fixed 'out-of-range' bug after 'second run') 2015-10-26: Version 16 (works with ImageJ 1.50d) 2014-07-18: Version 15 2014-02-03: Version 14 2012-11-22: First version |
| Source: |
Align_Fluor_Channels_17.txt A demo stack is available. Older versions |
| Description: |
Repairs displacement of fluorescence channels in bacterial images. Channel #1 (phase contrast) is used as reference. |
| Features |
|
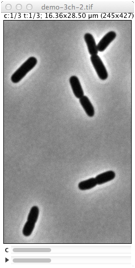
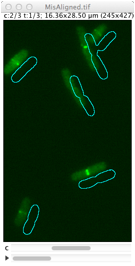
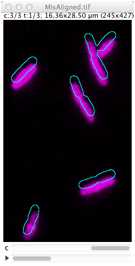
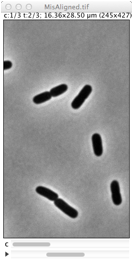
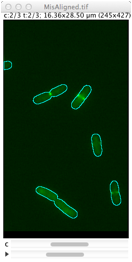
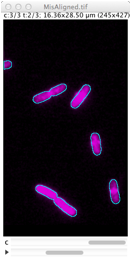
Derives cell outlines from channel 1 (PhC) and shows them in the current channel. This selection can manually be dragged so it matches the fluorescence pattern, but alignment is not performed yet. Use Edit> Select None (shift-A) to remove selection.
Shifts (translates) content of current channel by the amount the user has dragged the cell outlines
Removes current selection and overlay
You are asked to choose a folder containing hyperstacks. Then the misalignment of all images in that folder is calculated and stored in the same folder as table "Offsets.txt". Any old offsets file will be overwritten.
You are asked to choose a folder containing "Offsets.txt" with the corresponding stacks, which then will be aligned and saved. Note that the non-aligned images will be overwritten with the aligned ones, so make sure you have a back-up in case the result is not satisfactory. You can keep the Caps Lock down to observe or abort the alignment. For safety, "Offsets.txt" will be renamed to "AppliedOffset.txt" so it will not be used twice. In case of premature abortion, "Offsets.txt" may not be correct anymore.